Introduction
PRIME-Del is a precision genome deletion method based on prime editing. To facilitate prime editing gRNA (pegRNA) design used in PRIME-Del, we developed the Python-based script to generate pegRNA sequences. In this manual, we are going to introduce how to use the tool for paired pegRNA design.
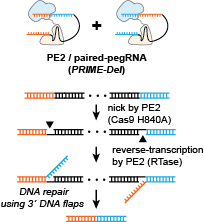
PRIME-Del takes a FASTA-formatted sequence file with a few parameters defining the desired deletion as the input. The user can either define deletion size or precise deletion location as parameters. We also highly recommend use scored gRNA files generated using Flashfry, CRISPOR or GPP sgRNA designer as input to identify effective CRISPR/Cas9 spacers. pegRNAs for deletions whose junctions do not fall at PAM sites can be designed using the option ‘--precise’ (-p), which adds insertion sequences to both pegRNAs to facilitate the desired edit. PegRNA sequences that include RNA polymerase III terminator sequences (more than four consecutive T’s) are filtered out. The software generates warning messages if more than 4 out of 5 bp in either 3’-DNA-flap are either G or C.
Using PRIME-Del
Webtool
We developted a webtool for quick design. It takes a sequence and a few paramaters as input. it will design all possible paired pegRNAs match the criteria and create a txt file for download. We recommend the input target sequence is 2x longer than the desired deletion size.
Install local PRIME-Del from Github
PRIME-Del has been wrapped as a python package. Code is available at Github
Requirements
Compatible with Python3.5 or higher, should work with Python2.7 (not tested).
Requires pandas,numpy, regex and xlrd
In terminal, type:
git clone https://github.com/shendurelab/Prime-del.git
cd Prime-del
pip install primedel-1.1-py2.py3-none-any.whl We provide a simple test python code gen_pegs.py for you to test your target region. It takes a FASTA file and a few parameters as input, and outputs the a file with pegRNA pairs match the design paramters.
python gen_pegs.py -i examples/eNMU.fasta -f examples/eNMU-flashfry.output.scored -x 500 1000 -p For more information, please check out our paper(Junhong Choi, Wei Chen, et al.)